This section provides different modules that support the weekly lessons, including Journal Club material and a detailed ‘How To’ guidance for EMOD.
Overview of EMOD and dtk simulation process
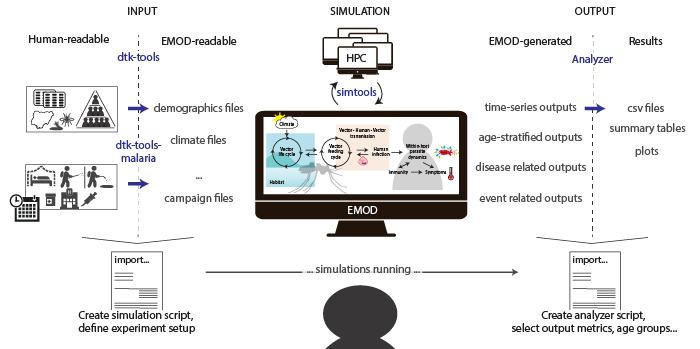
The figure above shows from the left to the right the process of preparing a simulation, over letting the simulation run, to analyzing model outputs to receive results.
INPUT: When preparing a simulation, input files readable in EMOD (json format) need to be generated from data, often in format of prepared input csv files. In this step a single python is used to import relevant methods (functions) from dtk-tools and dtk-tools-malaria that 1) handles the conversion to json for input files, 2) builds the simulation scenario files, and 3) submits the simulation to run automatically.
SIMULATION:
The simulations are send to run either locally [LOCAL], or on a cluster (on IDM’s owned cluster called COMPS [HPC], or on Northwestern University’s cluster QUEST [NUCLUSTER]). Running on a cluster allows to run many single simulations in parallel and is much faster than running locally. This step does not requrie any further action from the user, except for occasional checking on their status of completion.
OUTPUT: After the simulation finished running one or more output files were generated by EMOD that describe transmission and disease or event related outcomes over time, per age bin or other categories selected. The analyzer functions from dtk-tools and dtk-tools-malaria are used again via import in a single python script (’analyzer’) to obtain results in the format of csv files, summary tables or plots as specified.
simtools: For the process to run, dtk needs to know where to access the input files from and where to sent them to. The directories are specified in a specific file (simtools.ini) and in scripts initialized via SetupParser.init('HPC'). A simtools.ini file is required within the same directory where the scripts that uses dtk functions are located (simulation and analyzer scripts).
